
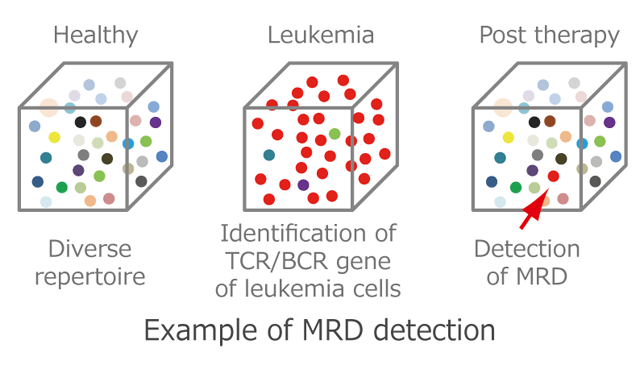
Observations in the setting of a non-defined tumor antigen (EBV − DLBCL) were contrasted with those with a well-defined tumor antigen (EBV + DLBCL) and with a non-lymphomatous cancer (melanoma) that is typically associated with a high mutational load. However here, particular attention has been paid to the association with progression-free and overall survival and TCR repertoire after conventional frontline (i.e., non-checkpoint blockade) therapy.
There have been several studies in solid cancers examining the dynamics of the TCR repertoire after treatment with immune checkpoint inhibitors. This is the first intra-tumoral TCR repertoire study of a large diffuse large B-cell lymphoma (DLBCL) cohort. However, the role of the intra-tumoral T-cell receptor (TCR) repertoire has not been established. Several studies have highlighted the importance of various aspects of the tumor microenvironment upon the clinical outcome in lymphoma. Highly dominant T-cell clonal expansions within the TME are associated with poor outcome in DLBCL treated with conventional frontline therapy. Increased T-cell diversity was associated with significantly elevated PD-1, PD-L1, and PD-L2 immune checkpoint molecules.Ĭonclusions: Put together, these findings suggest that the TCR repertoire is a key determinant of the TME. In keeping, clonal expansions were most pronounced in the EBV + DLBCL subtype that is known to express immunogenic viral antigens and is associated with particularly poor outcome. In DLBCL, a highly dominant single T-cell clone was associated with inferior 4-year OS rate of 60.0%, compared with 79.8% in patients with a low dominant clone (95% CI, 66.7%–88.5% P = 0.005). Results: The TCR repertoire within DLBCL nodes was abnormally narrow relative to non-diseased nodal tissues ( P < 0.0001). The primary endpoints were 4-year overall survival (OS) and progression-free survival (PFS). Key immune checkpoint genes within the TME were digitally quantified by nanoString. Purpose: To investigate the relationship between the intra-tumoral T-cell receptor (TCR) repertoire and the tumor microenvironment (TME) in de novo diffuse large B-cell lymphoma (DLBCL) and the impact of TCR on survival.Įxperimental Design: We performed high-throughput unbiased TCRβ sequencing on a population-based cohort of 92 patients with DLBCL treated with conventional (i.e., non-checkpoint blockade) frontline “R-CHOP” therapy.


 0 kommentar(er)
0 kommentar(er)
